Rutledge Group research
Specific interests include:
- peptides, proteins and enzyme mechanism
- metallo-enzymes and enzyme evolution
- metal sensing in vivo and in vitro
- target activation strategies for drug development
- catalyst development
- biosynthesis
- antibiotics and bacterial resistance
Our research uses the tools of organic synthesis, bio-organic and bio-inorganic chemistry to develop new biologically-inspired catalysts for important synthetic transformations and the cleanup of environmental contaminants. Nature uses exquisitely tuned enzyme systems to achieve a diverse array of complex chemical reactions, many of which currently have no counterpart in chemical synthesis. Our aim is to develop systems which mimic these enzymes and to use these, along with modified protein catalysts, as new reagents for difficult synthetic transformations, and in cell-free bioremediation processes to break down environmental pollutants.
Battling the Superbugs, New Antibiotics and New Strategies
The rise and rise of the so-called Superbugs is well documented in the media. Bacteria that are resistant to (i.e. not killed by) most current antibiotics are increasingly widespread, and the need for new drugs and new strategies to combat them grows ever more important. We are widely interested in antibiotics chemistry and antibiotic biosynthesis, and are pursuing a number of approaches to meet the challenges posed by antibiotic resistant bacteria.
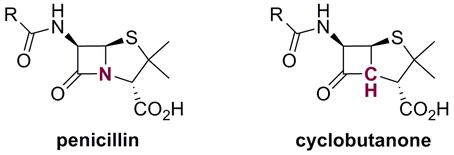
Current strategies include the synthesis and evaluation of new cyclobutanone antibiotics and the design of antibiotics with novel double-punch and resistance-activated modes of action.
Oxidising hydrocarbons: Iron for strength
The efficient and selective oxidation of hydrocarbon substrates is of interest to chemists and biologists alike. The conversion of simple hydrocarbons (alkanes, alkenes and aromatic compounds) to functionalised targets (alcohols, diols, epoxides and carbonyl compounds) is of great interest in synthetic and medicinal chemistry, but many of these key transformations are inaccessible with current synthetic methods. And the oxidative break down of polycyclic aromatic hydrocarbon pollutants (PAHs, toxic, carcinogenic contaminants present in high concentrations around various industrial sites) is an important goal in environmental rehabilitation.
Nature uses highly efficient enzyme systems to oxidise hydrocarbons in high yield with complete selectivity. We are working to develop new catalysts inspired by these biological systems, and to use them for the selective oxidation of hydrocarbon substrates: new reagents for synthesis and for the environment.
Enzymes from the non-heme iron(II) oxidase (NHIO) family catalyse a diverse range of oxidations in Nature, including the amino acid oxidations, conversion of naphthalene and toluene to diol products, and oxidative steps in the biosynthesis of penicillin and cephalosporin antibiotics.
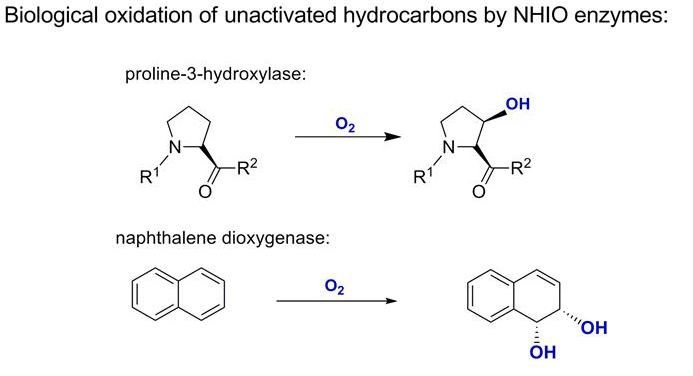
The X-ray crystal structures of various NHIO enzymes show that the active site region is highly conserved across the whole family: the non-heme iron(II) centre is bound by two histidine residues and a single carboxylic acid side chain from the protein, a motif appropriately called the 2-His-1-carboxylate facial triad. This key, oxygen-activating motif involves iron(II) ligated by two nitrogen ligands and two or three oxygen ligands.
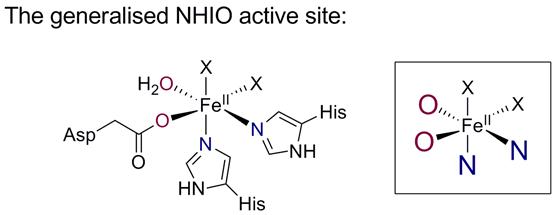
We have synthesised a series of small-molecule systems that mimic this active site and are developing these complexes as efficient catalysts for biomimetic hydrocarbon oxidation and iron-mediated C�H activation.
In collaboration with Dr Nick Coleman in the School of Life and Environmental Sciences at the University of Sydney, we are investigating new methods for biocatalytic hydrocarbon oxidation, developing whole-cell mycobacterial systems as the ultimate green chemistry reagents for oxidising alkene and alkane substrates.
Mercury Rising: Sensors for Heavy Metals
Heavy metals like cadmium and mercury form some of the most toxic materials known. Both these elements attack their target organs at very low concentrations, the kidneys are most affected, while cadmium also attacks the lungs and mercury the nervous system. Various industrial activities have caused levels of these heavy metals in the environment to rise over recent years, particularly in areas that have seen intensive mining activities and industrial sites such as incinerators.
Some plants and microbes have developed strategies to live in locales prone to high levels of cadmium and mercury pollution, using sulfur-rich proteins such as metallothioneins and phytochelatins to bind and the toxic metal ions and sequester them away from the rest of the cell's machinery. These proteins incorporate a very high percentage of cysteine residues ~ ca. 30% of the primary structure. It is this characteristic that makes them effective agents for heavy metal sequestration: the numerous sulfur atoms bind mercury and cadmium very tightly indeed. Our approach builds on that seen in Nature: we are using thiol- and sulfide-rich peptides as agents for sensing and binding mercury and cadmium in the environment.
One Crazy Active Site: Nitrile hydratase Mimics and Mechanism
Cyanide and organic nitriles are well known chemical toxins, but such compounds also have considerable commercial utility and are used in a range of industrial processes. Significant quantities of nitrile-containing waste are currently dumped at sea or pumped into deep pressure wells, despite their carcinogenicity and the threat to the environment. Nitrile hydratase enzymes play central roles in the breakdown of nitriles in vivo, and are also used as biocatalysts in the industrial synthesis of polymers and fine chemicals incorporating the amide functional group.
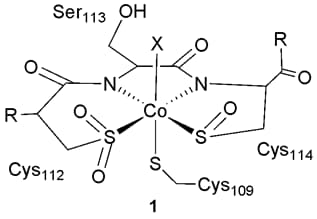
Crystal structures have been solved for nitrile hydratases from several organisms, and they show a remarkable ligand environment around the active site metal (1), which is either iron(III) or cobalt(III). Two nitrogen atoms from main-chain amides bind to the metal, along with three sulfur atoms, each in a different oxidation state! It is also remarkable that these five metal binding ligands are located in a very short section of the protein's primary sequence: they are spread across only six amino acids in one peptide chain.
We are working to develop new peptide-based systems as bio-inspired catalysts for nitrile hydration. We also wish to study the unusual sulfur oxidation that occurs at the nitrile hydratase active site, and to elucidate a detailed mechanism for nitrile hydratase catalysis.