Past projects
A clearer look at galaxies with probabilistic modelling
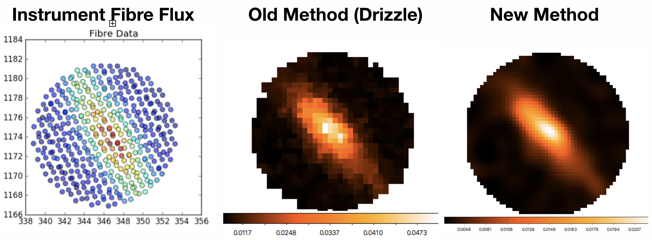
Working with Professor Scott Croom, Dr Richard Scalzo and the SAMI team, we developed a novel method applying Bayesian probability to optimally fuse images from a spectral fibre bundle, providing uniform image quality while maintaining spectral details.
The SAMI Galaxy Survey is a large-scale program to observe several thousand galaxies with the University of Sydney-built Sydney-AAO Multi-object Integral field spectrograph (SAMI). It is challenging to reconstruct an image from the telescope’s optical fibres, which capture 500 spectra at several locations across a galaxy, and the previous method produced several undesirable artefacts.
Working with Professor Scott Croom from the School of Physics, Dr Richard Scalzo from the Centre for Translational Data Science, and the SAMI team, we developed a novel method applying Bayesian probability to optimally fuse images from a spectral fibre bundle, providing uniform image quality while maintaining spectral details. This innovative technology is implemented as flexible software framework that can eventually be deployed at a wide range of worldwide telescopes. While our software begins to help astrophysicists study the universe, publications deriving from this work are in preparation.
Towards AI in mobile health for heart disease

By applying techniques from natural language processing, predictive machine learning and descriptive data analysis, we curated outgoing messages and presented insights into how participants engage in mobile health.
Cardiovascular disease is one of the highest causes of death in Australia, and chest pain the second most common presentation at Australian emergency departments. Westmead Applied Research Centre has been developing text messaging interventions to improve lifestyle factors associated with cardiovascular risk. Towards scaling up these trials and applying artificial intelligence to personalise the interventions, Professor Clara Chow and Dr Harry Klimis sought our insights into the data from past clinical trials.
By applying techniques from natural language processing, predictive machine learning and descriptive data analysis, we curated their outgoing messages and presented insights into how participants engage in mobile health. We helped them use content analysis techniques to manually categorise their data, and our advice on applying artificial intelligence for personalisation helped secure a $1M prize from the Google Impact AI Challenge.
Saving Australian endangered animals from extinction

Bioinformaticians at the Sydney Informatics Hub have supported the Australasian Wildlife Genomics Group, led by Professor Kathy Belov in the conservation of Australian wildlife.
The Australasian Wildlife Genomics Group, led by Professor Kathy Belov, lead research programmes in the conservation of Australian wildlife. This includes creating a library of digital genomes of 40 endangered Australian animals (Koala genome published in Nature, 2018 DOI) and developing therapies for contagious facial cancers in Tasmanian devils. They are also exploring therapies against superbugs in humans using naturally occurring antibiotics from the pouch of Australian marsupials.
Bioinformaticians at the Sydney Informatics Hub have supported the Belov lab by processing next generation sequencing data through SIH high-throughput transcriptome and variant calling pipelines, and assisted with the generation of linked-read de novo genome assemblies and scaffolding draft assemblies with Hi-C data. We have also supported access and facilitated use of National HPC facilities and Commercial Cloud services. The Belov lab are part of the recently awarded Centre of Excellence in Innovations in Peptide and Protein Science and are supported by ARC Discovery Project and Linkage Project grants.
Discoveries driven by the DNA of ancient tribes

Photo courtesy of Chris Bennett
Bioinformaticians at the Sydney Informatics Hub have supported the Haye’s lab by processing whole genome sequencing data through SIH high-throughput germline and somatic variant calling pipelines. We have also supported access to, and facilitated use of, National HPC facilities.
The Haye’s Lab, led by Professor Vanessa Hayes, explores the complex genetics of indigenous southern African people to gain insights into the origins of humanity and in therapies for prostate cancer. Professor Hayes and Dr Chan led a landmark study into the birthplace and first migrations of modern humans (Nature, 2019 DOI). The Haye’s Lab are also investigating biomarkers that can predict the risk of prostate cancer. The Hayes lab are supported by Philanthropic funding and NHMRC Project Grants.
Bioinformaticians at the Sydney Informatics Hub have supported the Haye’s lab by processing whole genome sequencing data through SIH high-throughput germline and somatic variant calling pipelines. We have also supported access to, and facilitated use of, National HPC facilities.
Video tracking predator-prey interactions in fish
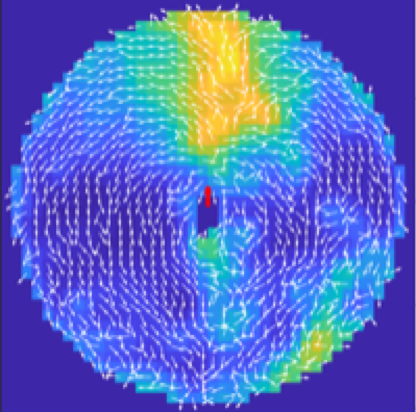
By video-tracking the interaction between prey and their predator under controlled conditions, we provided fine-scale characterisation of how prey adapt their behaviour according to their continuous assessment of risk.
By video-tracking the interaction between prey mosquitofish, Gambusia holbrooki, and their predator, jade perch, Scortum barcoo, under controlled conditions, we provide some of the first fine-scale characterisation of how prey adapt their behaviour according to their continuous assessment of risk based on both predator behaviour and angular distance to the predator’s mouth. When these predators were inactive and posed less of an immediate threat, prey were often found within the attack cone of the predator showing reductions in speed and acceleration, characteristic of predator-inspection behaviour. However, when predators became active, prey swam faster with greater acceleration and were closer together within the attack cone of predators.
Most importantly, this study provides evidence that prey do not adopt a uniform response to the presence of a predator. Instead, we demonstrate that prey are capable of rapidly and dynamically updating their assessment of risk and showing fine-scale adjustments to their behaviour.
The statistician, Dr Gordon McDonald, assisted Mia Kent and Prof Ashley Ward from the Faculty of Science, by creating data visualisations and time series analyses.
The work was published in this paper: “Fine-scale behavioural adjustments of prey on a continuum of risk”. M.I.A. Kent, J.E. Herbert-Read, G.D. McDonald, A.J. Wood, A.J.W. Ward. Proceedings of the Royal Society B. 2019
Restriction of anthropogenic foods alters a top predator’s diet and intra-specific interactions
CC Image courtesy of Dwayne Madden on Flickr
Our statistical consulting team provided expertise in data analysis and visualisation to explore effects of anthropogenic foods to a top predator's diet and intra-specific interactions.
Research outcome
- Altering anthropogenic food sources impacts dingo’s dietary preferences, as well as inter and intra species dynamics.
- There was evidence for an increase in feral cat predation by the dingo in areas where anthropogenic foods were removed. Efforts to manage food waste could therefore have positive flow on effects to natural eco-systems through reduced predation of high risk and endangered species by the feral cat.
How statistical consulting contributed
General discussion and advice on issues such as:
- Identification of a Quasi BACI design which strengthens the evidence for causation (as opposed to correlation).
- Initial Exploratory Data Analysis to identify potential issues and clarify the experimental design.
- General Linear Model with Poisson distribution, with standard assumption testing as well as the not so standard (zero inflation, over dispersion).
- Visualisations that quantify both the impact and also allow for direct evaluation of null hypothesis using p-values.
Researcher quote
"The assistance I got from SIH helped to inform the study design and analysis. As a result the project was publishable in a quality journal and the outcomes were more relevant to managers on the ground. I now send all students in my lab to get assistance from SIH before starting any field study” – Dr Thomas Newsome